Accessing Planetary Computer STAC files in DuckDB
I’ve been informed that this method is no longer working, and I haven’t had time to try and update it to work again. I’m leaving this here for historical value, and in case anyone can adapt the method to make it work.
Microsoft Planetary Computer is a wonderful archive of geospatial datasets (primarily raster images of various types), provided with a STAC catalog to enable them to be easily searched through an API. That’s fine for normal usage where you want to find a selection of images and access the images themselves, but less useful when you want to do some analysis of the metadata itself.
For that use case, Planetary Computer provide the STAC metadata in bulk, stored as GeoParquet files. Their documentation page explains how to use this with geopandas and do some large-ish scale processing with Dask. I wanted to try this with DuckDB – a newer tool that is excellent at accessing and processing Parquet files, including efficiently accessing files available via HTTP, and only downloading the relevant parts of the file. So, this post explains how I managed to do this – showing various different approaches I tried and how (or if!) each of them worked.
Getting URLs
Planetary Computer URLs are basically just URLs to files on Azure Blob Storage. However, the URLs require signing with a Shared Access Signature (SAS) key. Luckily, there is a Planetary Computer Python module that will use an API to generate the correct keys for us.
In the docs for using the STAC GeoParquet files, they give this code:
catalog = pystac_client.Client.open(
"https://planetarycomputer.microsoft.com/api/stac/v1/",
modifier=planetary_computer.sign_inplace,
)
asset = catalog.get_collection("io-lulc-9-class").assets["geoparquet-items"]This gets a collection, and the geoparquet-items collection-level asset, having used a ‘modifier’ to sign the URLs as they are acquired from the STAC API. The URL is then stored in asset.href.
Using the DuckDB CLI – with HTTP URLs
When using GeoPandas, the docs show that you can pass a storage_options parameter to the read_parquet method and it will ‘just work’:
df = geopandas.read_parquet(
asset.href, storage_options=asset.extra_fields["table:storage_options"]
)However, to use the file in DuckDB we need a full URL. The SQL code we’re going to use in DuckDB for a quick test looks like this:
SELECT * FROM <URL> LIMIT 1This just gets the first row of whatever URL is given.
Unfortunately, though, the URL provided by the STAC catalog looks like this:
abfs://items/io-lulc-9-class.parquetIf we try that with DuckDB, we find it doesn’t know how to deal with it. So, we need to convert this to a standard HTTP URL that can be downloaded as if it were just typed into a web browser (or used with cURL or wget). After a bit of playing around, I found I could create a full URL with this Python code:
url = f'https://{asset.extra_fields["table:storage_options"]
["account_name"]}.dfs.core.windows.net/{asset.href[7:]}?
{asset.extra_fields["table:storage_options"]["credential"]}'That’s taking the account_name and sticking it in as part of the host part of the UK, then extracting the path from the original URL and adding that, and then finishing with the SAS token (stored as credential in the storage options) as a URL query parameter.
That results in a URL that looks like this (split over multiple lines for readability):
https://pcstacitems.dfs.core.windows.net/items/io-lulc-9-class.parquet
?st=2024-06-14T19%3A04%3A20Z&se=2024-06-15T19%3A49%3A20Z&sp=rl&sv=2024-05-04&
sr=c&skoid=9c8ff44a-6a2c-4dfb-b298-1c9212f64d9a
&sktid=72f988bf-86f1-41af-91ab-2d7cd011db47&skt=2024-06-15T09%3A00%3A14Z
&ske=2024-06-22T09%3A00%3A14Z&sks=b
&skv=2024-05-04
&sig=A0e%2BihbAagHmIZ%2Bg8gzH71TavRYQMZiHWJ/Uk9j0Who%3D(note: that specific URL won’t work for you, as the SAS tokens are time-limited).
If we insert that into our DuckDB SQL statement then we find it works (click to enlarge):
Great! Now let’s try with a different collection on Planetary Computer – in this case the Sentinel 2 collection. We can make the URL in the same way as before, by changing how we define asset:
asset = catalog.get_collection("sentinel-2-l2a").assets["geoparquet-items"]and then using the same query in DuckDB with the new URL. Unfortunately, this time we get an error:
Invalid Input Error: File '<URL>' too small to be a Parquet fileThe problem here is that for larger collections, the STAC metadata is spread out over a series of GeoParquet files inside a folder, and the URL we’ve been given is just to the folder (even though it ends in .parquet). As we’re just using HTTP, there is no way to do things like list the files in a folder, so DuckDB has no way to find out what files are within the folder and start reading them. We need to find another way.
Using the DuckDB CLI – with the Azure extension
Conveniently, there is an Azure extension for DuckDB, and it lets you use URLs like 'abfss://⟨my_filesystem⟩/⟨path⟩/⟨my_file⟩.⟨parquet_or_csv⟩'. That’s a slightly different URL scheme to the one we’ve been given (abfss as opposed to abfs), but we can easily sort that.
Looking at the authentication docs though, it seems to require you to specify either a connection string, a service principal or using the ‘Azure Credential Chain’ (which, I think, works with the az command line and various environment variables that you may have set up). We don’t have any of those – they’re all a far broader scope than what we’ve got, which is just a SAS token for a specific file or folder. It looks like the Azure extension doesn’t support this, so we’ll have to look for another way.
Using the DuckDB Python library – with an Azure connector
As well as the command-line interface, DuckDB can also be used from Python. To set this up, just run pip install duckdb. While you’re at it, you might want to install pandas and the adlfs library for connecting to Azure Storage.
Using this is actually quite simple, first import the libraries:
import duckdb
from adlfs.spec import AzureBlobFileSystemthen set up the Azure connection:
a = AzureBlobFileSystem(account_name='pcstacitems',
sas_token=asset.extra_fields["table:storage_options"]['credential'])Note how here we’re using the sas_token parameter to provide a SAS token. We could use a different parameter here to provide a connection string or some other credential type. I couldn’t find much real-world use of this sas_token parameter when looking online – so this is probably the key ‘less well documented’ bit to take away from this article.
Continuing, we then connect to DuckDB and ‘register’ this Azure connection:
connection = duckdb.connect()
connection.register_filesystem(a)From there, we can run a SQL query like this:
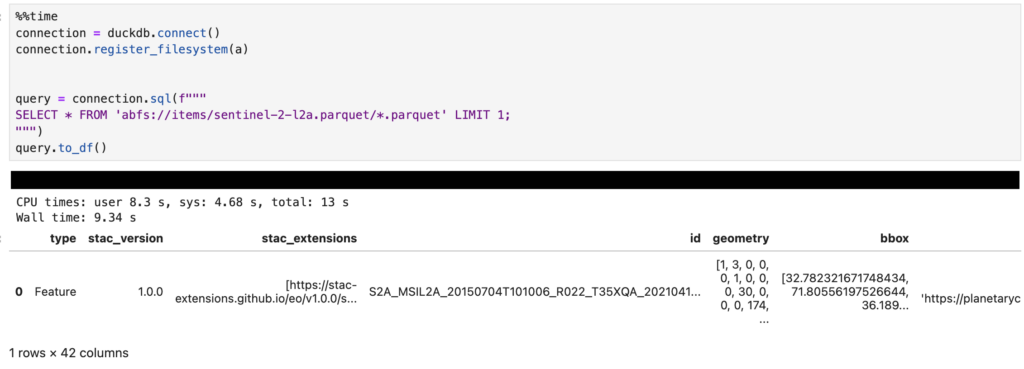
query = connection.sql(f"""
SELECT * FROM 'abfs://items/sentinel-2-l2a.parquet/*.parquet' LIMIT 1;
""")
query.to_df()The call to to_df at the end converts the result into a Pandas DataFrame for easy viewing and manipulation. The results are shown below (click to enlarge):
Note that we’re passing a URL of abfs://items/sentinel-2-l2a.parquet/*.parquet – this is the URL from the STAC catalog with /*.parquet added to the end to ensure DuckDB picks up the large number of Parquet files stored there. I’d have thought this would have worked without that, but if I miss that out I get a error saying:
TypeError: int() argument must be a string, a bytes-like object or a real number, not 'NoneType'I suspect this is something to do with how things are passed to the Azure connection that we registered with DuckDB, but I’m not entirely sure. If you do know, then please leave a comment!
A few fun queries
So, now we’ve got this working, what can we do? Well, we can – fairly efficiently – run queries across all the STAC metadata for Sentinel 2 images. I’ll just give a few examples of queries I’ve run below.
We can find out how many images there are in the collection in total:
SELECT COUNT(*) FROM 'abfs://items/sentinel-2-l2a.parquet/*.parquet'We can do the same for just the images acquired in 2020 – by using a fact we know about how the individual parquet files are named (from the Planetary Computer docs):
SELECT COUNT(*) FROM 'abfs://items/sentinel-2-l2a.parquet/*2020*.parquet'And we can start looking at how many scenes were captured with different amounts of cloud cover:
SELECT COUNT(*) / (SELECT COUNT(*) FROM 'abfs://items/sentinel-2-l2a.parquet/*2020*.parquet'), CASE
WHEN "eo:cloud_cover" between 0 and 5 then '0-5%'
WHEN "eo:cloud_cover" between 5 and 10 then '5-10%'
WHEN "eo:cloud_cover" between 10 and 15 then '10-15%'
END AS category
FROM 'abfs://items/sentinel-2-l2a.parquet/*2020*.parquet' GROUP BY category,This tells us that 20% of scenes had between 0 and 5% cloud cover (quite a high number, I thought – but then again, I am used to living in the UK!), and around 4-5% in each of the 5-10% and 10-15% categories.
There are plenty of other analyses that you could do with these Parquet files, of course. At some point I might even get around to the task which initially made me look into this: that is, trying to find Landsat and Sentinel 2 scenes that were acquired over the same location at very similar times. I think I’ll leave that for another day though…
If you found this post useful, please consider buying me a coffee.
This post originally appeared on Robin's Blog.
Categorised as: Academic, GIS, How To, Python, Remote Sensing

Leave a Reply